Publications
See Google Scholar
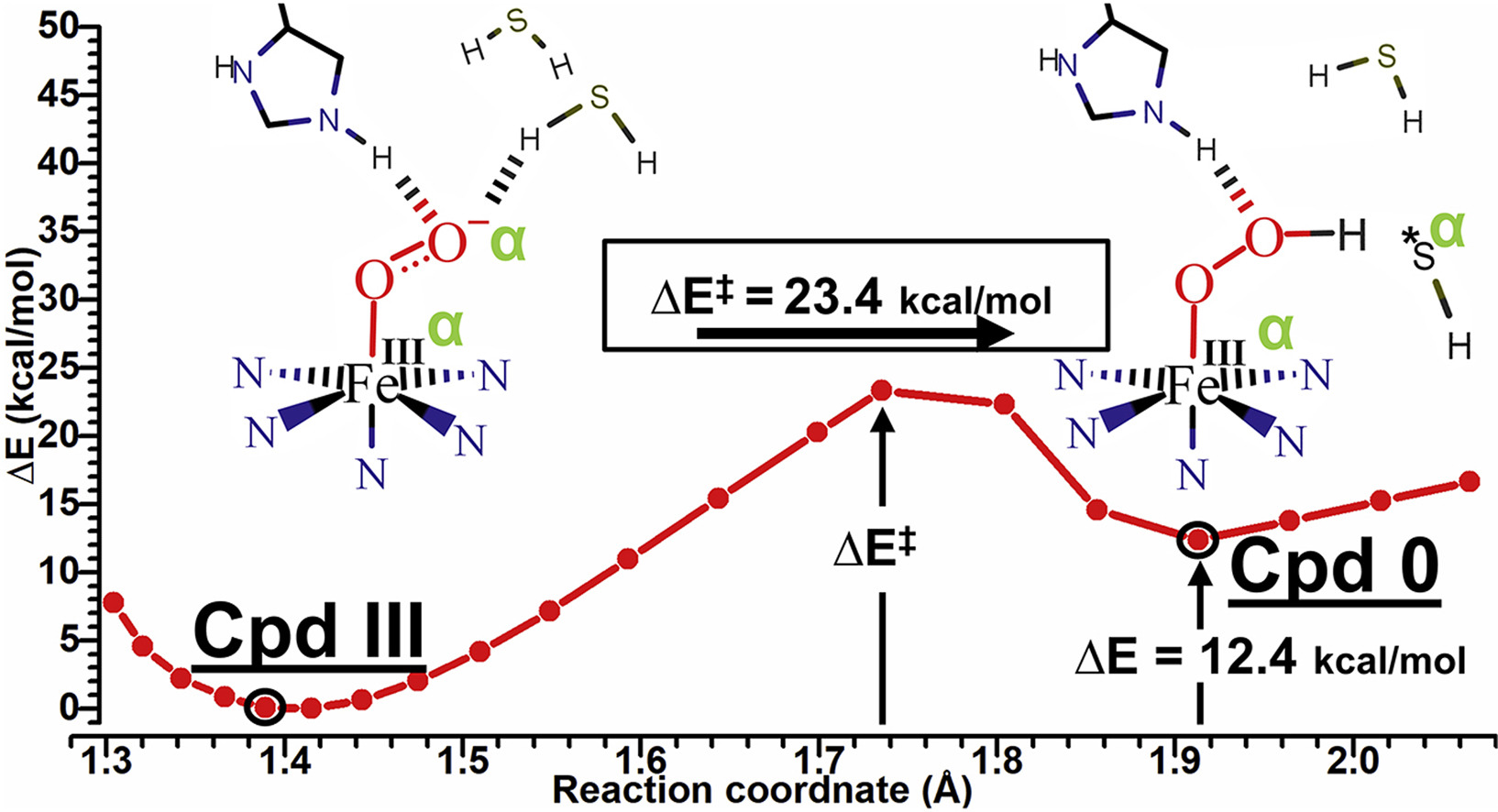
A Reaction Pathway to Compound 0 Intermediates in oxy-Myoglobin through Interactions with Hydrogen Sulfide and His64
Angel D. Rodriguez-Mackenzie Hector D. Arbelo-Lopez, Troy Wymore, Juan Lopez-Garriga
Journal of Molecular Graphics and Modeling, 2020, 94:107465.
doi: 10.1016/j.jmgm.2019.107465
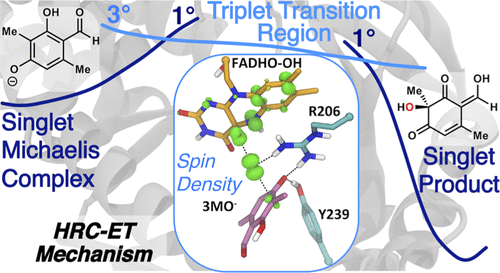
The Hydroxyl Radical Coupled Electron Transfer Mechanism of Flavin-Dependent Hydroxylases
Sara E. Tweedy, Attabey Rodríguez Benítez, Alison R. H. Narayan, Paul M. Zimmerman, Charles L. Brooks III and Troy Wymore*
Journal of Physical Chemistry B, 2019, 123(38):8065-8073.
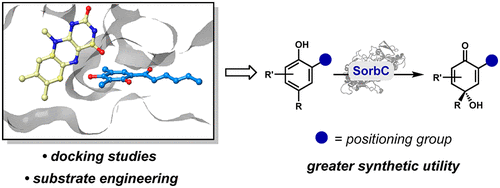
Positioning-Group-Enabled Biocatalytic Oxidative Dearomatization
Summer A. Baker Dockrey, Carolyn Suh, Attabey Rodríguez Benítez, Troy Wymore, Charles L. Brooks III and Alison R. H. Narayan
ACS Central Science, 2019, 5(6):1010-1016.
doi: 10.1021/acscentsci.9b00163
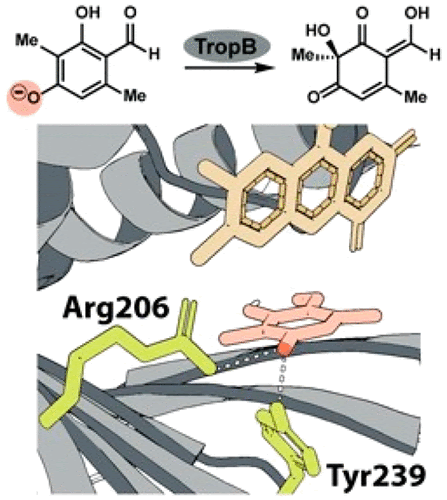
Structural Basis for Selectivity in Flavin-dependent Monooxygenase-catalyzed Oxidative Dearomatization
Attabey Rodríguez Benítez, S Tweedy, SA Baker Dockrey, AL Lukowski, T Wymore, D Khare, CL Brooks III, BA. Palfey, JL Smith, Alison R. H. Narayan
ACS Catalysis, 2019, 9(4):3633-3640.

Neutron Scattering in the Biological Sciences: Progress and Prospects
One of Several Authors
Acta Crystallography D Struct Biol, 2018, 9(4):3633-3640.
doi: 10.1107/S2059798318017503
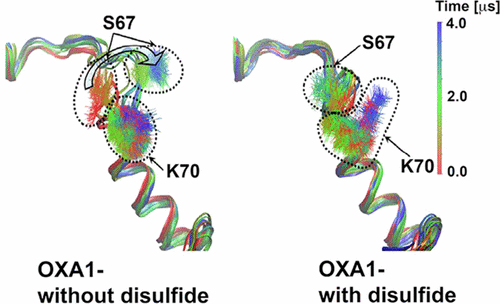
A Distal Disulfide Bridge in OXA-1 β-Lactamase Stabilizes the Catalytic Center and Alters the Dynamics of the Specificity Determining Ω Loop
N. Simakov, DA Leonard, JC Smith, T. Wymore, A. Szarecka
Journal of Physical Chemistry B, 2017, 121:3285-3296.
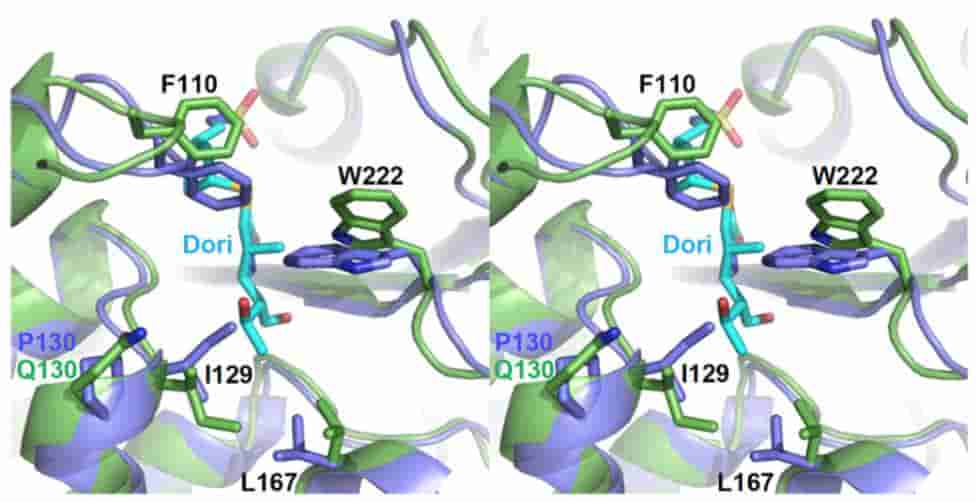
Clinical Variants of the Native Class D β-Lactamase of Acinetobacter baumannii Pose an Emerging Threat through Increased Hydrolytic Activity against Carbapenems
EC Schroder, ZL Klamer, A Saral, KA Sugg, CM June, T. Wymore, A Szarecka, DA Leonard
Antimicrob Agents Chemother, 2016, 60:6155-6164.
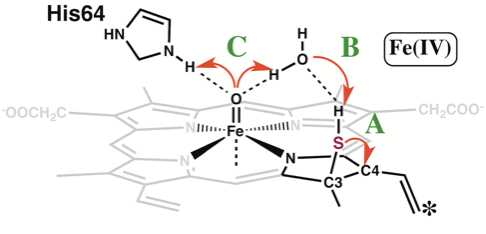
Homolytic Cleavage of Both Heme-Bound Hydrogen Peroxide and Hydrogen Sulfide Leads to the Formation of Sulfheme
H. Arbelo, N. Simakov, J. C. Smith, Juan Lopez-Garriga and T. Wymore
Journal of Physical Chemistry B, 2016, 120:7319-7331.
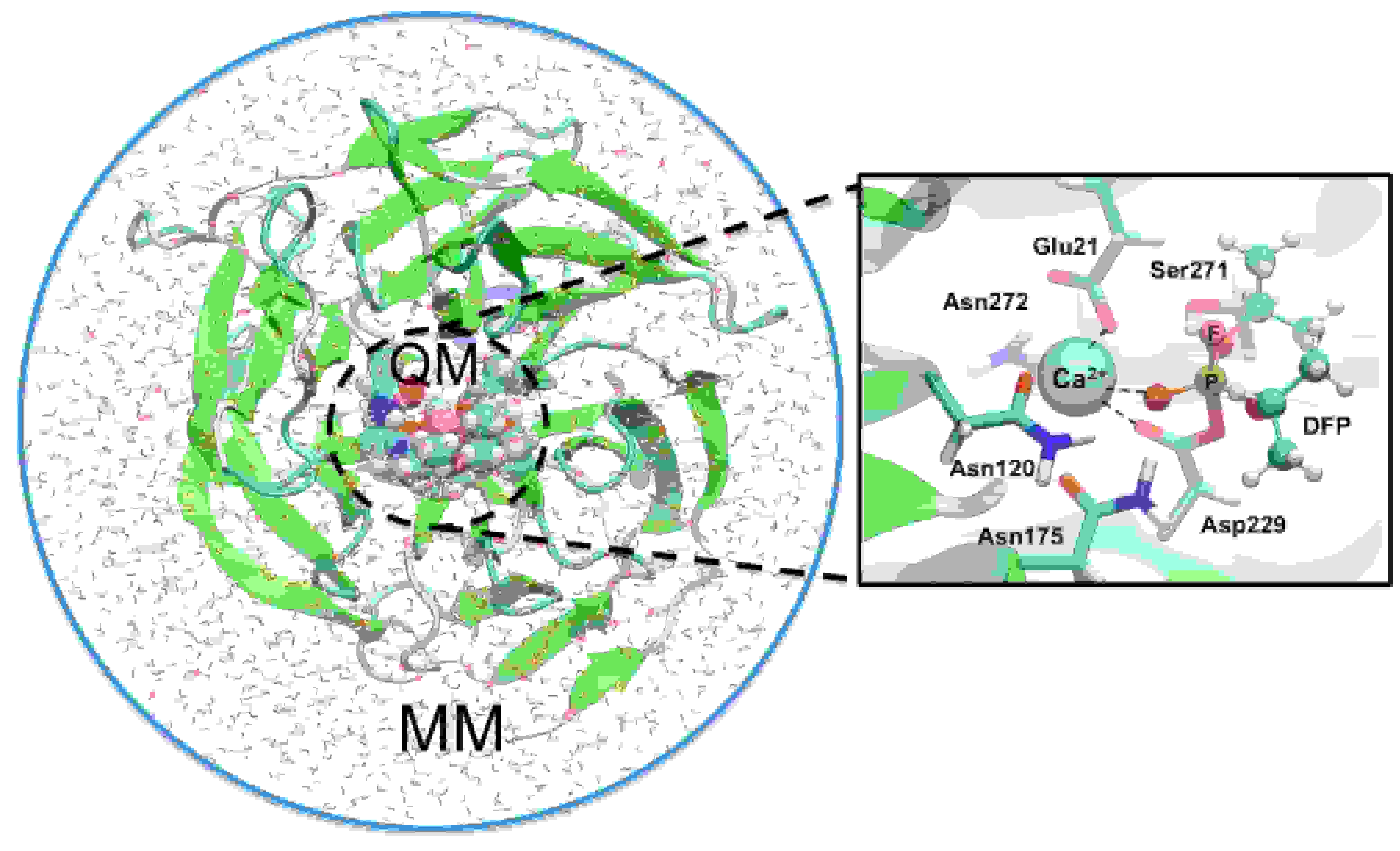
Multiscale Modeling of Nerve Agent Hydrolysis Mechanisms: A Tale of Two Nobel Prizes
M. J. Field and T. Wymore
Invited Review, Physica Scripta, 2014, 89:108004 (11 pages).
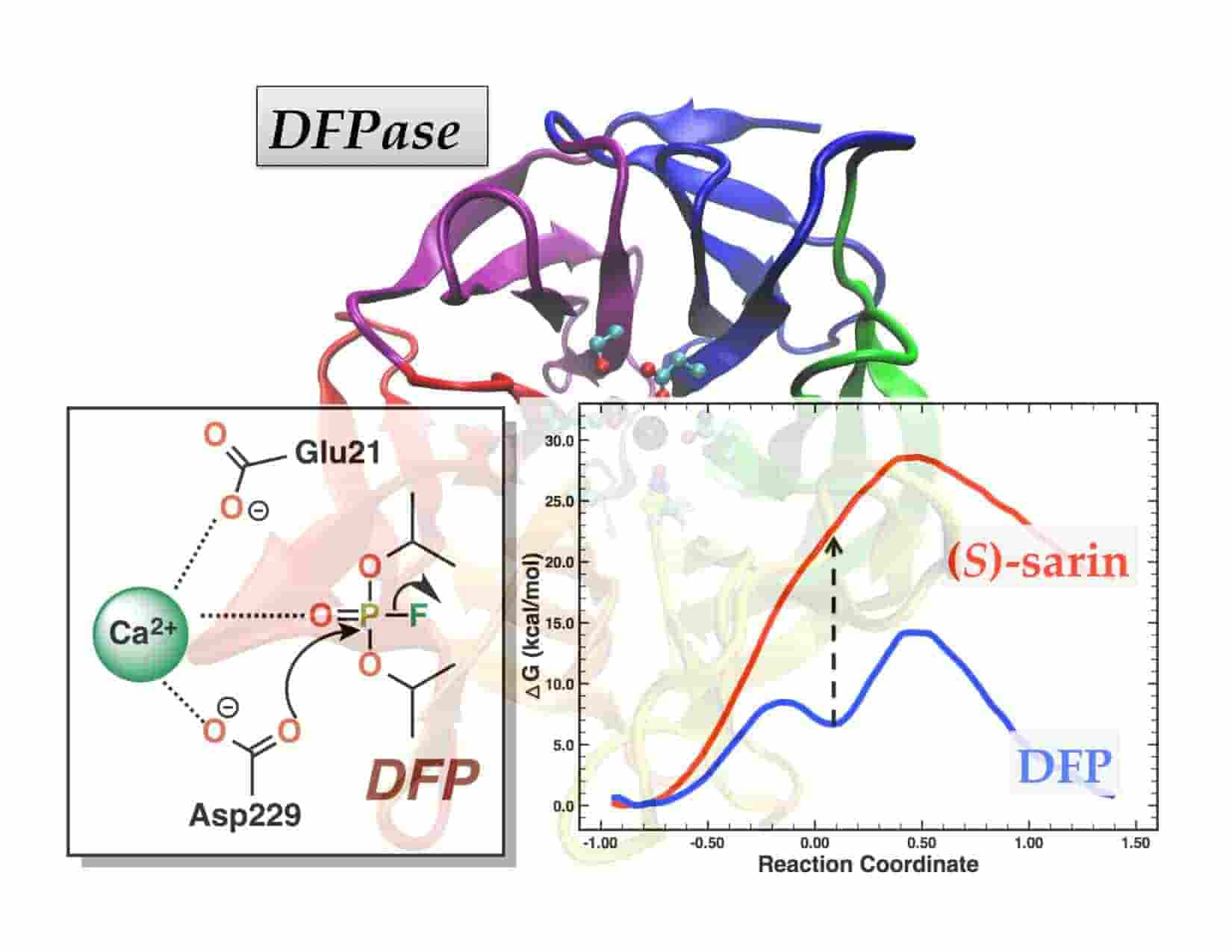
Hydrolysis of DFP and (S)-Sarin by DFPase proceeds along two different reaction pathways:Implications for Engineering Bioscavengers
T. Wymore*, M. J. Field, P. Langan, J. C. Smith, and J. M. Parks
Journal of Physical Chemistry B, 2014, 118:4479-89.
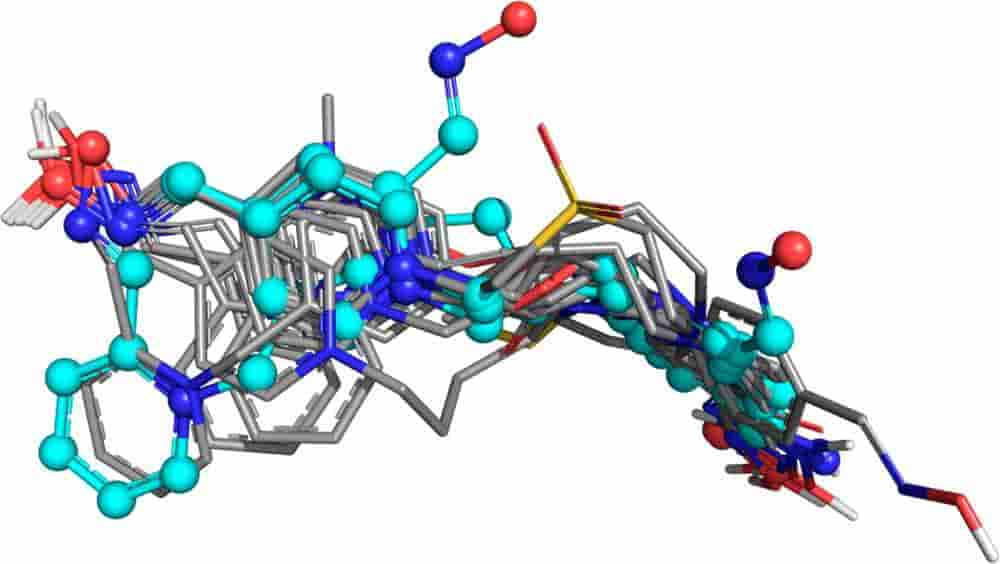
Exploring the Physicochemical Properties of Oxime-Reactivation Therapeutics for Cyclosarin, Sarin, Tabun, and VX Inactivated Acetylcholinesterase
E. X. Esposito, T. R. Stouch, T. Wymore and J. D. Madura
Chemical Research in Toxicology, 2014, 27:99-110.
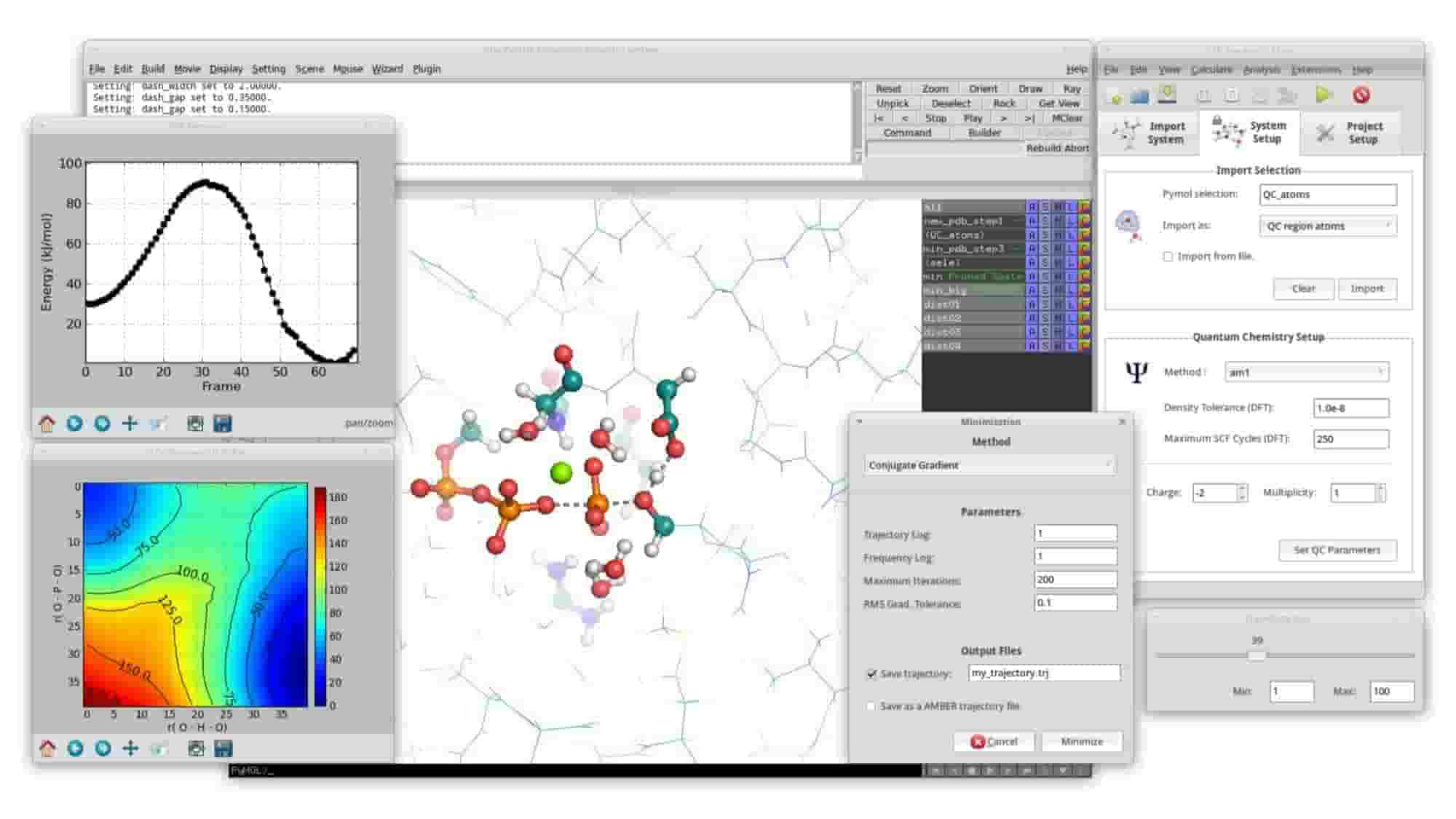
GTKDynamo: a PyMOL plug-in for QC/MM Hybrid Potential Simulations.
J. Fernando Bachega, L. F. Timmers, L. Assirati, L. R. Bachega, M. J. Field, T. Wymore.
Journal of Computational Chemistry, 2013, 34:2190-2196.
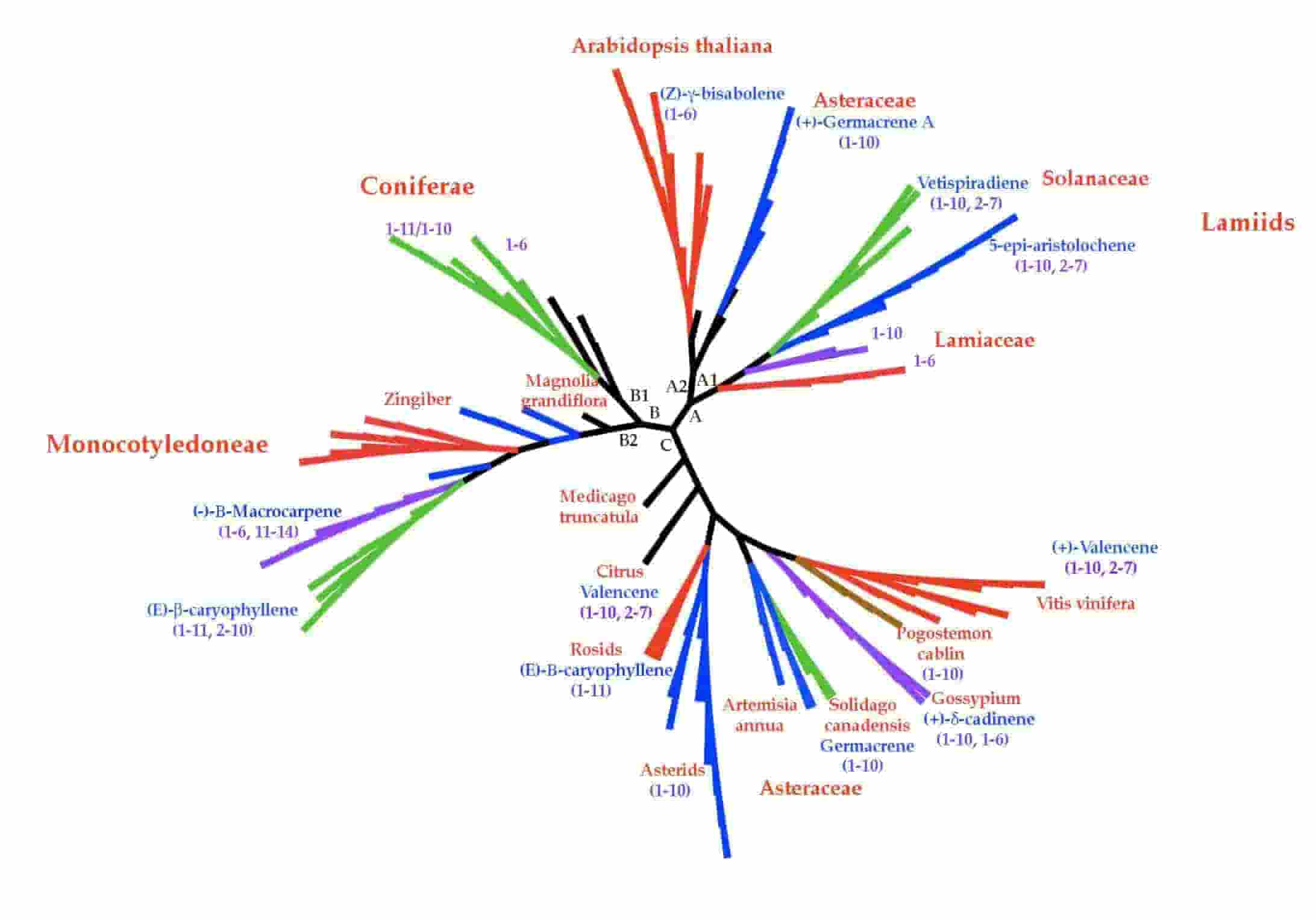
A Mechanism for Evolving Novel Plant Sesquiterpene Synthase Function
T. Wymore*, B. Y. Chen, H. B. Nicholas Jr., A. J. Ropelewski and C. L. Brooks III
Molecular Informatics, 2011, 30:896-906.
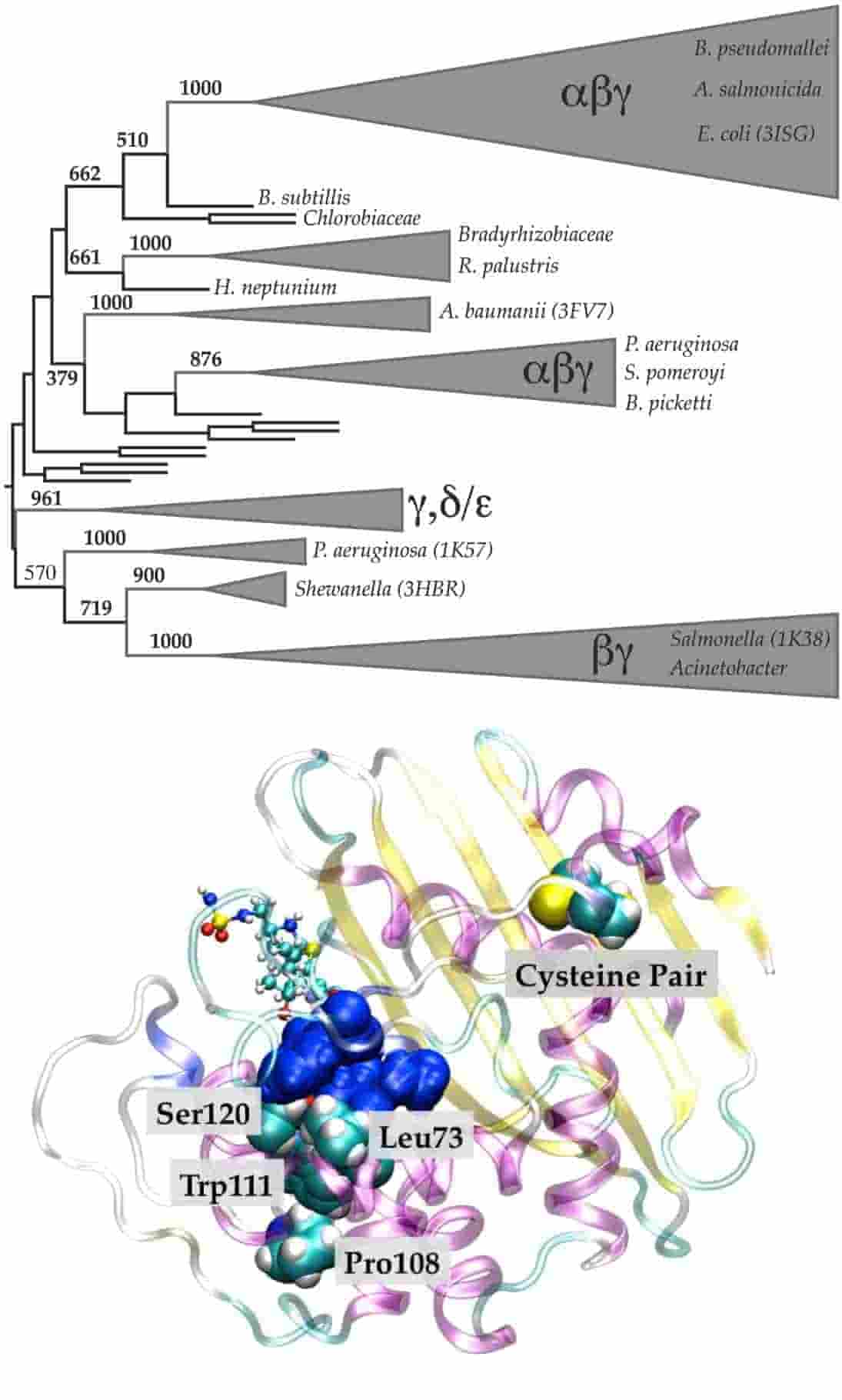
The class D Beta-lactamase Family: Residues Governing the Maintenance and Diversity of Function
A. Szarecka, K. R. Lesnock, C. Ramirez-Mondragon, B. Y. Chen, H. B. Nicholas Jr. and T. Wymore*
Protein Engineering, Design and Selection, 2011, 24:801-809.
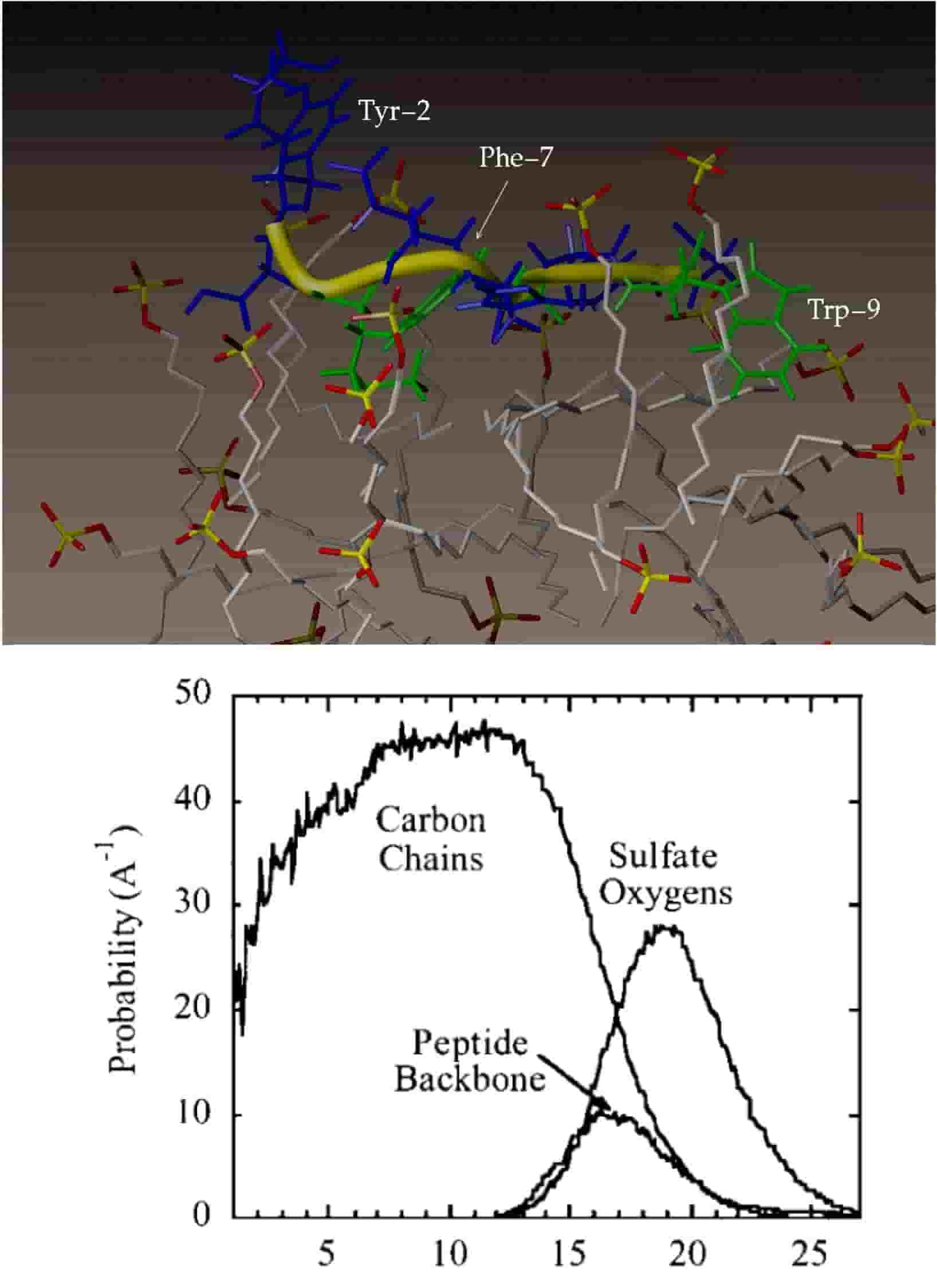
Structure and Dynamics of ACTH(1-10) on the Surface of the Sodium Dodecylsulfate (SDS) Micelle: A Molecular Dynamics Simulation Study
T. Wymore and T. C. Wong
Journal of Biomolecular Structure and Dynamics, 2000, 18:461-476.
doi: 10.1080/07391102.2000.10506681
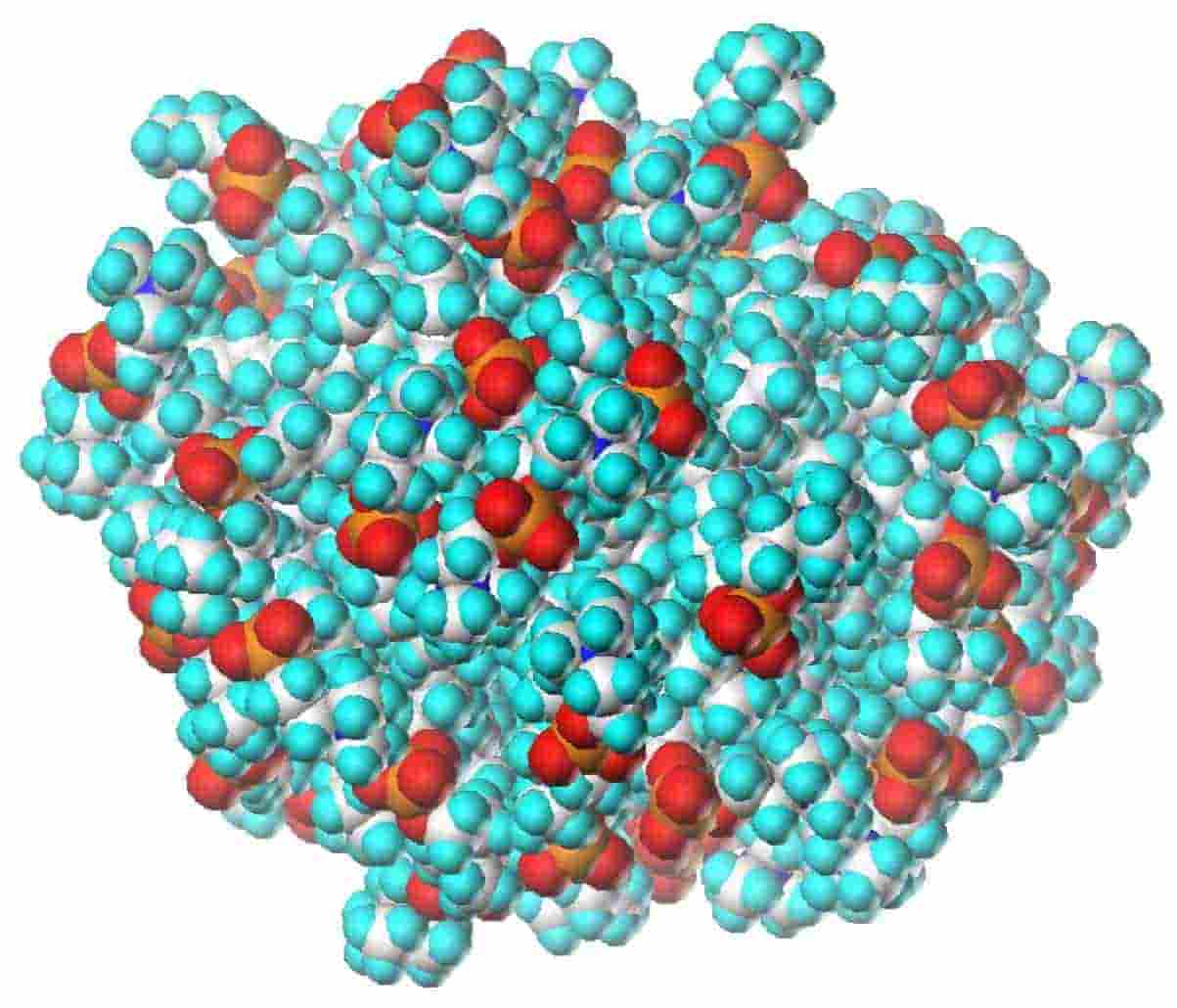
Molecular Dynamics Simulation of the Structure and Dynamics of a Dodecylphosphocholine Micelle in Aqueous Solution
T. Wymore, X. Gao and T. C. Wong
Journal of Molecular Structure, 1999, 485-6:195-210.
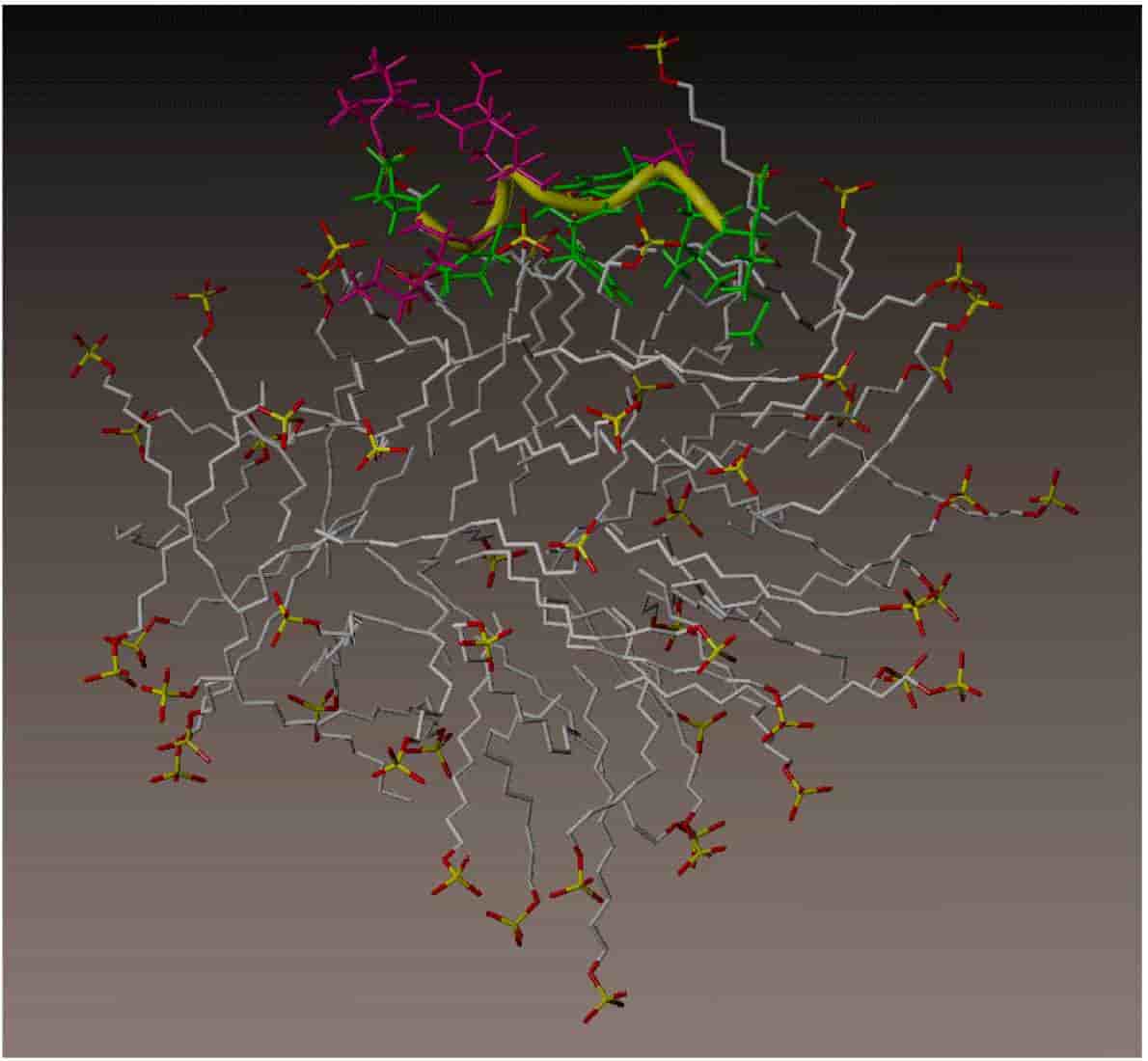
Molecular Dynamics Study of Substance P Peptides Partitioned in an SDS Micelle
T. Wymore and T. C. Wong
Biophysical Journal, 1999, 76:1213-1227.
doi: 10.1016%2FS0006-3495(99)77285-1
Molecular Dynamics Study of Substance P Peptides in a Biphasic Membrane Mimetic
T. Wymore and T. C. Wong
Biophysical Journal, 1999, 76:1199-1212.
doi: 10.1016%2FS0006-3495(99)77284-X
Full List
A Reaction Pathway to Compound 0 Intermediates in oxy-Myoglobin through Interactions with Hydrogen Sulfide and His64
Angel D. Rodriguez-Mackenzie Hector D. Arbelo-Lopez, Troy Wymore, Juan Lopez-Garriga
Journal of Molecular Graphics and Modeling, 2020, 94:107465.
doi: 10.1016/j.jmgm.2019.107465
The Hydroxyl Radical Coupled Electron Transfer Mechanism of Flavin-Dependent Hydroxylases
Sara E. Tweedy, Attabey Rodríguez Benítez, Alison R. H. Narayan, Paul M. Zimmerman, Charles L. Brooks III and Troy Wymore*
Journal of Physical Chemistry B, 2019, 123(38):8065-8073.
doi: 10.1021/acs.jpcb.9b08178
Room-temperature X-ray structures of human acetylcholinesterase capture VX-induced reorientation of oxime reactivator and reveal binding of a VX hydrolysis product
O. Gerlits, M. Fajer, X. Cheng, T. Wymore, D. Blumenthal, P. Taylor, Z. Radić, A. Kovalevsky
Journal of Biological Chemistry, 2019, 294(27):10607-10618.
doi: 10.1074/jbc.RA119.008725
Positioning-Group-Enabled Biocatalytic Oxidative Dearomatization
Summer A. Baker Dockrey, Carolyn Suh, Attabey Rodríguez Benítez, Troy Wymore, Charles L. Brooks III and Alison R. H. Narayan
ACS Central Science, 2019, 5(6):1010-1016.
doi: 10.1021/acscentsci.9b00163
Structural Basis for Selectivity in Flavin-dependent Monooxygenase-catalyzed Oxidative Dearomatization
Attabey Rodríguez Benítez, S Tweedy, SA Baker Dockrey, AL Lukowski, T Wymore, D Khare, CL Brooks III, BA. Palfey, JL Smith, Alison R. H. Narayan
ACS Catalysis, 2019, 9(4):3633-3640.
doi: 10.1021/acscatal.8b04575
Neutron Scattering in the Biological Sciences: Progress and Prospects
One of Several Authors
Acta Crystallography D Struct Biol, 2018, 9(4):3633-3640.
doi: 10.1107/S2059798318017503
Charge Transfer and π to π* Transitions in the Visible Spectra of Sulfheme Met Isomeric Structures
Arbelo-López, HD, Rodriguez-Mackenzie, AD, Roman-Morales, EM, Wymore, T and López-Garriga, J
Journal of Physical Chemistry B, 2018, 122(19):4947-4955.
doi: 10.1021/acs.jpcb.7b12393
A Distal Disulfide Bridge in OXA-1 β-Lactamase Stabilizes the Catalytic Center and Alters the Dynamics of the Specificity Determining Ω Loop
N. Simakov, DA Leonard, JC Smith, T. Wymore, A. Szarecka
Journal of Physical Chemistry B, 2017, 121:3285-3296.
doi: 10.1021/acs.jpcb.6b07884
Clinical Variants of the Native Class D β-Lactamase of Acinetobacter baumannii Pose an Emerging Threat through Increased Hydrolytic Activity against Carbapenems
EC Schroder, ZL Klamer, A Saral, KA Sugg, CM June, T. Wymore, A Szarecka, DA Leonard
Antimicrob Agents Chemother, 2016, 60:6155-6164.
doi: 10.1128/AAC.01277-16
Homolytic Cleavage of Both Heme-Bound Hydrogen Peroxide and Hydrogen Sulfide Leads to the Formation of Sulfheme
H. Arbelo, N. Simakov, J. C. Smith, Juan Lopez-Garriga and T. Wymore
Journal of Physical Chemistry B, 2016, 120:7319-7331.
doi: 10.1021/acs.jpcb.6b02839
Long-Range Electrostatics-Induced Proton Transfer in a Protein Visualized by Neutron Crystallography
O. Gerlits, T. Wymore, A. Das, Chen-Hsiang Shen, J. M. Parks, J. C. Smith, K. Weiss, D. A. Keen, M. P. Blakeley, P. Langan, I. Weber, A. Kovalevsky.
Angewandte Chemie, 2016, 55:4924-4927.
doi: 10.1002/anie.201509989
The Structural Basis of Activity Against Aztreonam and Enhanced Spectrum Cephalosporins for Two Carbapenem-Hydrolyzing Class D β-Lactamases from Acinetobacter Baumannii
J.M. Mitchell, J. R. Clasman, C. M. June, K-C. J. Kaitany, J. R. LaFleur, M. A. Taracila, N. V. Klinger, R. A. Bonomo, T. Wymore, A. Szarecka, R. A. Powers and D. A. Leonard
Biochemistry, 2015, 54:1976-1987.
doi: 10.1021/bi501547k
Multiscale Modeling of Nerve Agent Hydrolysis Mechanisms: A Tale of Two Nobel Prizes
M. J. Field and T. Wymore
Invited Review, Physica Scripta, 2014, 89:108004 (11 pages).
doi: 10.1021/jp410422c
L-Arabinose Binding, Isomerization, and Epimerization by D-Xylose Isomerase: X-Ray/Neutron Crystallographic and Molecular Simulation Study
P. Langan, A. K. Sangha, T. Wymore, J. Parks, Z. K. Yang, B. L. Hanson, Z. Fisher, S. Mason, M. P. Blakely, T. Forsyth, J. P. Glusker, H. L. Carrell, J. C. Smith, D. A. Keen, D. E. Graham and A. Kovalevsky
Structure, 2014, 22:1287-1300.
doi: 10.1016/j.str.2014.07.002
Hydrolysis of DFP and (S)-Sarin by DFPase proceeds along two different reaction pathways:Implications for Engineering Bioscavengers
T. Wymore*, M. J. Field, P. Langan, J. C. Smith, and J. M. Parks
Journal of Physical Chemistry B, 2014, 118:4479-89.
doi: 10.1021/jp410422c
Exploring the Physicochemical Properties of Oxime-Reactivation Therapeutics for Cyclosarin, Sarin, Tabun, and VX Inactivated Acetylcholinesterase
E. X. Esposito, T. R. Stouch, T. Wymore and J. D. Madura
Chemical Research in Toxicology, 2014, 27:99-110.
doi: 10.1021/tx400350b
GTKDynamo: a PyMOL plug-in for QC/MM Hybrid Potential Simulations.
J. Fernando Bachega, L. F. Timmers, L. Assirati, L. R. Bachega, M. J. Field, T. Wymore.
Journal of Computational Chemistry, 2013, 34:2190-2196.
doi: 10.1002/jcc.23346
Asymmetric Ligand Binding Facilitates Conformational Transitions in Pentameric Ligand-gated Ion Channels
M. H. Cheng, D. Mowrey, L. T. Liu, D. Willenbring, T. Wymore, Y. Xu and P. Tang.
Journal of American Chemical Society, 2013, 135:2172-2180.
doi: 10.1021/ja307275v
From Molecular Phylogenetics to Quantum Chemistry: Discovering Nature’s Design Principles through
Computation
C. L. Brooks III and T. Wymore*
Invited Review for Special Issue, Computational and Structural Biotechnology Journal, 2012, 2:e201209018.
doi: 10.5936/csbj.201209018
A Mechanism for Evolving Novel Plant Sesquiterpene Synthase Function
T. Wymore*, B. Y. Chen, H. B. Nicholas Jr., A. J. Ropelewski and C. L. Brooks III
Molecular Informatics, 2011, 30:896-906.
doi: 10.1002/minf.201100087
The class D Beta-lactamase Family: Residues Governing the Maintenance and Diversity of Function
A. Szarecka, K. R. Lesnock, C. Ramirez-Mondragon, B. Y. Chen, H. B. Nicholas Jr. and T. Wymore*
Protein Engineering, Design and Selection, 2011, 24:801-809.
doi: 10.1093/protein/gzr041
The Plasmodium Glutathione S-Transferase: Bioinformatics Characterization and Classification into the Sigma
Class
E. Colon-Lorenzo, A. Serrano, H. B. Nicholas, T. Wymore, A. Ropelewski and R. Gonzalez-Mendez
Proceedings of Bioinformatics, 2010, 1st International Conference on Bioinformatics, 173-180. Lisbon, Portugal, ISBN: 978-989-674-019-1.
experts.umich.edu
Gamma-glutamyl semialdehyde dehydrogenase: simulations on native and mutant forms support the importance
of outer shell lysines
J. Hempel, A. Kraut and T. Wymore
Chemico-Biological Interactions, 2009, 178(1-3):75- 78.
doi: 10.1016/j.cbi.2008.10.009
Mechanistic Implications of the Cysteine-Nicotinamide Adduct in Aldehyde Dehydrogenase Based on Quantum
Mechanical/Molecular Mechanical Simulations
T. Wymore*, D. W. Deerfield II and J. Hempel
Biochemistry, 2007, 46: 9495-9506.
doi: 10.1021/bi700555g
A Quantum Chemical Study of the Mechanism of Action of Vitamin K Epoxide Reductase (VKOR): II.
Transition States
C. H. Davis, D. W. Deerfield II, T. Wymore, D. W. Stafford, L. G. Pedersen
J. Molecular Graphics and Modeling, 2007, 26:401-408.
doi: 10.1016/j.jmgm.2006.10.005
A Quantum Chemical Study of the Mechanism of Action of Vitamin K Carboxylase (VKC): III. Intermediates
and Transition States
C. H. Davis, D. W. Deerfield II, T. Wymore, D. W. Stafford, L. G. Pedersen
J. Molecular Graphics and Modeling, 2007, 26:409-414.
doi: 10.1016/j.jmgm.2006.10.006
Unexpected Encounters in Simulations of the ALDH Mechanism
J. Hempel, H. B. Nicholas Jr., S. T. Brown, T. Wymore
Enzymology and Molecular Biology of Carbonyl Metabolism, 2007, 13:9-13 Weiner, H., B. Plapp, R.Lindahl, and E. Maser, eds. Purdue University Press, Lafayette, IN.
doi: 10.1021/bi700555g
The Use of Alternative Alignments and Structural Scoring Methods for Comparative Protein Structure Modeling
A. C. Marko, K. Stafford and T. Wymore*
Journal of Chemical Information and Modeling, 2007, 47:1263-1270.
doi: 10.1007/978-1-60761-842-3_6
Quantum Chemical Study of the Mechanism of Action of Vitamin K Epoxide Reductase (VKOR)
D. W. Deerfield II, C. H. Davis, T. Wymore, D. W. Stafford, L. G. Pedersen
Int. Journal of Quantum Chemistry, 2006, 106:2944-2952.
doi: 10.1016/j.jmgm.2006.10.005
Molecular Recognition of Aldehydes by Aldehyde Dehydrogenase and Mechanism of Nucleophile Activation
T. Wymore*, J. Hempel, S. S. Cho, A. D. MacKerell, H. B. Nicholas and D. W. Deerfield II
PROTEINS: Struct. Func. Bioinf, 2004, 57:758-771 (COVER Article)..
doi: 10.1002/prot.20256
Parametrization of 2-Bromo- 2-Chloro- 1,1,1-Trifluoroethane (Halothane) and Hexafluoroethane for Nonbonded
Interactions
Z. Liu, Yan Xu, A. C. Saladino, T. Wymore, and P. Tang
Journal of Physical Chemistry A, 2004, 108(5):781-786.
doi: 10.1021/jp0368482
Initial Events in class 3 Aldehyde Dehydrogenase: MM and QM/MM simulations
T. Wymore*, D. W. Deerfield II, M. J. Field, H. B. Nicholas Jr. and J. Hempel
Chemico-Biological Interactions, 2003, 143-144:75-84.
doi: 10.1016/S0009-2797(02)00175-8
An Algorithm for Identification and Ranking of Family-Specific Residues, Applied to the ALDH3 family
J. Hempel, J. Perozich, T. Wymore and H. B. Nicholas Jr.
Chemico-Biological Interactions, 2003, 143-144:23-28.
doi: 10.1016/S0009-2797(02)00165-5
Molecular Dynamics Simulation of class 3 Aldehyde Dehydrogenase
T. Wymore*, H. B. Nicholas and J. Hempel
Chemico-Biological Interactions, 2001, 130-132:201-207.
doi: 10.1021/bi962734n
Structure and Dynamics of ACTH(1-10) on the Surface of the Sodium Dodecylsulfate (SDS) Micelle: A Molecular
Dynamics Simulation Study
T. Wymore and T. C. Wong
Journal of Biomolecular Structure and Dynamics, 2000, 18:461-476.
doi: 10.1080/07391102.2000.10506681
Molecular Dynamics Simulation of the Structure and Dynamics of a Dodecylphosphocholine Micelle in Aqueous
Solution
T. Wymore, X. Gao and T. C. Wong
Journal of Molecular Structure, 1999, 485-6:195-210.
doi: 10.1021/jp001268f
Molecular Dynamics Study of Substance P Peptides Partitioned in an SDS Micelle
T. Wymore and T. C. Wong
Biophysical Journal, 1999, 76:1213-1227.
doi: 10.1016%2FS0006-3495(99)77285-1
Molecular Dynamics Study of Substance P Peptides in a Biphasic Membrane Mimetic
T. Wymore and T. C. Wong
Biophysical Journal, 1999, 76:1199-1212.
doi: 10.1016%2FS0006-3495(99)77284-X
Adverse Effects on Plasma Polymerized Films due to Oxygen Etching in an Electrodeless Reactor
T. Wymore and M. F. Nichols
J. Vac. Sci. Technol. A, 1994, 12, 677-680.
doi: 10.1116/1.578852